Research

AI-powered Smart Microscopy for Stem-Cell Engineering
Cell engineering is predicted to be part of a regenerative medicine focused future. The human Mesenchymal Stem Cells (hMSC) are a cell type of major interest that are available from bone marrow, adipose tissue, and umbilical cord sources. Over 900 hMSC clinical trials have been conducted since 2004 and thousands of academic publications on potential hMSC treatments are published yearly. Despite this significant investigative effort, only few therapeutic products containing hMSCs or their derivative cells are approved yearly, mainly due to hMSC therapies requiring extensive safety testing by an independent contract research organization, sterile cold chain, and increased surgical time.
Our goal is to introduce a completely new paradigm in stem cell culturing where the standard trial and error approach to identify a suitable recipe is substituted by quantitative and continuous optimization. We intend to implement a next-generation bio-manufacturing process in which the stimulation is finely tuned in real time, based on the specific dynamics of the population, to overcome the issues associated with biological diversity and patient-specific conditions. The possibilities to combine different imaging technologies, to trigger imaging modalities based on the real-time image analysis results, and to synchronize image acquisition with external devices for sample manipulation (i.e., liquid handling or cell manipulation) enable the automation of complex workflows. For more info about how to join, please click here.
Image credits: Amanda Kwieraga

Deep neural network automated segmentation of cellular structures in volume electron microscopy
Three-dimensional electron-microscopy is an important imaging modality in contemporary cell biology. Identification of intracellular structures is laborious and time-consuming, however, and seriously impairs effective use of a potentially powerful tool. Resolving this bottleneck is therefore a critical next step in frontier biomedical imaging. We describe Automated Segmentation of intracellular substructures in Electron Microscopy (ASEM), a new pipeline to train a convolutional network to detect structures of wide range in size and complexity. We obtain for each structure a dedicated model based on a small number of sparsely annotated ground truth annotations from only one or two cells. To improve model generalization to developed a rapid, computationally effective strategy to refine an already trained model by including a few additional annotations. We show the successful automated identification of mitochondria, Golgi apparatus, endoplasmic reticulum, nuclear pore complexes, clathrin coated pits and coated vesicles, and caveolae in cells imaged by focused ion beam scanning electron microscopy with quasi-isotropic resolution. For more info click here.
Quantitative Absorption Microscopy – A Circulatory System on Chip
This project carried on at Massachusetts General Hospital, in collaboration with Ethan Schonbrun and John Higgins, is dedicated to the development of material technologies for mimicking the environment of the Circulatory System on Chip. In addition to the organ-on-chip fabrication, I am interested in performing quantitative measurements of other physical characteristics of cells, such as volume, protein mass, and oxygen tension. Resolving the distribution of different measured parameters enables better understanding of not only mean values of a cell population but also subtle differences in the response of each cell to its environment, genetic differences, and exposure to small molecules.
Image credit: Ethan Schonbrun
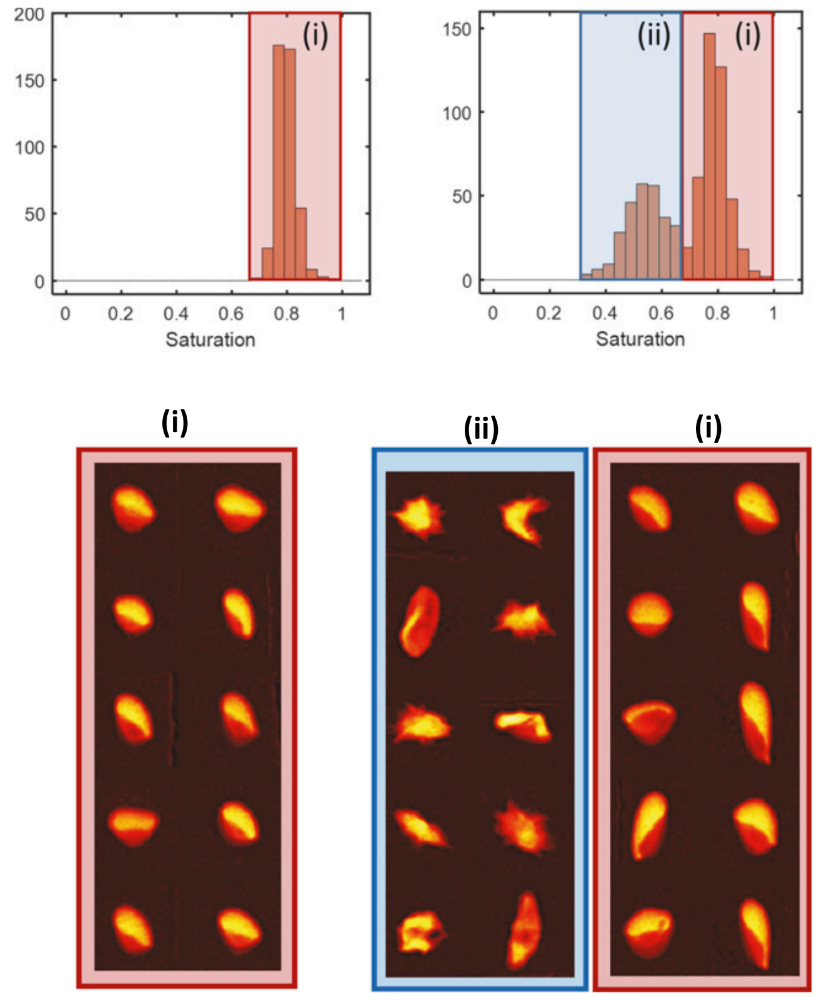
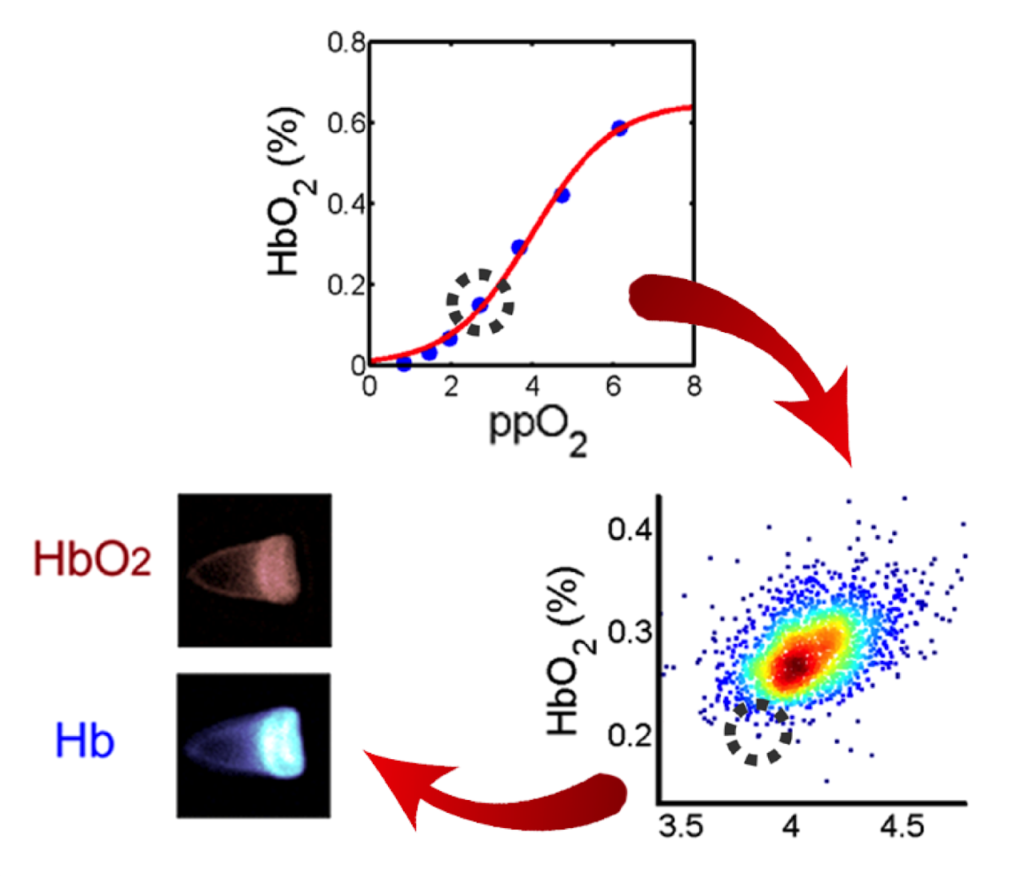
Sickle cell disease is the first “molecular disease” described more than 100 years ago. All morbidity and mortality are ultimately caused by polymerization of the variant sickle hemoglobin molecule in red blood cells (RBCs). However, despite decades of intense research, we are unable to measure the amount of hemoglobin polymer in individual RBCs with high throughput at arbitrary oxygen tension. We describe a high-throughput method to assess hemoglobin polymer at the single-cell level under controlled oxygen tension. The assay infers hemoglobin polymer fraction by measuring its effect on cellular oxygen affinity. This method provides biomarkers promising to improve management of sickle cell patients and to optimize the development and prioritization of candidate cures. For more info click here. This method provides biomarkers promising to improve management of sickle cell patients and to optimize the development and prioritization of candidate cures, and it’s currently used to validate the efficacy of BCL11A as therapeutic target in Sickle Cell Disease, Clinical Trials Identifier: NCT03282656 (Sponsor: David Williams – Boston Children’s Hospital).